Author Guidelines for Supplementary Materials in Optica Journals
Optica journals allow authors to include supplementary materials as integral parts of a manuscript. Such materials are subject to peer-review procedures along with the rest of the paper and should be uploaded and described using Optica's Prism manuscript system.
Supplementary Materials Supported in Optica Journals
Supplementary materials uploaded through Prism will be hosted in the OSA figshare portal. The material in figshare will not be publically accessible during peer review. Datasets and code may be uploaded to the Optica figshare portal or be placed in an appropriate external repository. See guidelines below.
Small data file such as data underlying a plot in a figure
Dataset stored in an appropriate external repository
Code or simulation files stored in an appropriate external repository
PDF document with expanded descriptions or methods (Optica only)
*Optica allows authors to include a supplemental document alongside letters and research articles that contains additional text, equations, citations, etc. (see Supplementary Materials in Optica for details). For all other Optica journals, supplemental text must be included as appendices within the primary manuscript.
1. Visualizations
Data visualizations typically illustrate a synopsis of research results. They are integral and as such should be:
- included only when they convey essential information beyond what can be presented within the article's PDF representation;
- peer reviewed along with the manuscript.
Image data that are appropriate for the paper but cannot be uploaded to OSA following the guidelines below can be treated as a Dataset as defined in this document.
Supported File Types
- (1) 2D
Supplementary two-dimensional (2D) image files should be included only if they provide critical information that cannot be conveyed adequately as a figure in a PDF page. 2D images should be submitted in JPEG, TIFF, or EPS format.
For example, a very-high-resolution biomedical image intended for image analysis would be appropriate as a supplementary 2D visualization whereas an image, graph, or chart that could be well represented as a figure in the manuscript generally would not be.
- (2) 3D
Supplementary 3D images should be submitted as a U3D (Universal 3D) object embedded in a PDF file viewable in Adobe Acrobat. Each 3D image should be placed in a separate PDF file that will be linked to the manuscript (typically as a link in a figure caption).
- (3) Video
Video files must use open compression standards for display on broadly available applications such as VLC or Windows Media Player. MOV, AVI, MPG, and MP4 video containers are accepted.
The following video guidelines will help with the submission process
- 15 MB is the recommended maximum video file size.
- 720 x 480 pixels (width by height) is the recommended screen size.
- If appropriate, insert a representative frame from the video in the manuscript as a figure.
- Minimize file size by using an acceptable codec such as x264 or XviD. HandBrake is an open source tool for converting video to common codecs.
- Videos must be playable on all platforms using VLC.
- Animations must be formatted into a standard video container.
Storage Location
OSA figshare platform.
How to Include in the Manuscript
Must be associated with a figure, table, or equation OR be referenced in the results section of the manuscript. Use the label "Visualization" and the item number to identify the visualization.
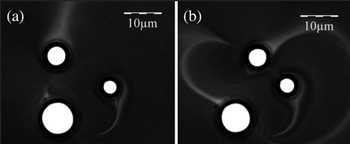
Fig. 5. Three traps create three rings of magnetic nanoparticles. The rings interact with one another (see Visualization 3). [From Masajada et al., Opt. Lett. 38, 3910 (2013).]
2. Data File
Machine-readable numerical data can be useful for showing the "underlying data" in a figure image or for showing expanded information for a static table presented in a paper.
They are integral and as such should be:
- included only when they convey essential information beyond what can be presented within the article's PDF representation;
- peer reviewed along with the manuscript.
Machine-readable data that are appropriate for the paper but cannot be uploaded to OSA following the guidelines below can be treated as a Dataset as defined in this document.
Supported File Types
CSV (comma-separated values)
Spreadsheet applications such as MS Excel can read and save to CSV format. To the extent possible, CSV files should be self-contained. For example, they should include meaningful column heads and provide explanatory notes as appropriate in a comments section at the top of the file. CSV files may not contain binary information such as images or equation objects and should not exceed the row, column, and character limits supported by MS Excel (for example, 65,536 rows by 256 columns) or other popular spreadsheet applications.
Sample CSV file with comments. Each file should be self-contained and use the .csv file extension.
# Comment 1
# Comment 2
Header1, Header2, Header3
A, B, C
X, Y, Z
Storage Location
OSA figshare platform
How to Include in the Manuscript
Must be associated with a figure, table, or equation OR be referenced in the results section of the manuscript. Use the label "Data File" and the item number to identify the visualization.
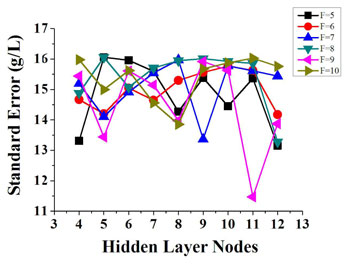
Fig. 4. Relationship among principal component, number of hidden layer nodes and standard deviation of correction set. F refers to principal component factors. See Data File 1 for underlying values. [From Ding et al., Biomed. Opt. Express 5, 1145 (2014).]
3. Citations to Large Datasets
Original contributions of large datasets should be placed in an open, archival database and cited in the main reference section. To ensure that readers are aware of the materials, the first mentions should use the following convention for providing a label and item number in addition to the citation: ". . . as we show in Dataset 1 (Ref. [2])."
Large datasets should be included as integral material that is peer reviewed along with the manuscript. Datasets can be uploaded to the OSA figshare platform or be placed in an appropriate external repository.
Consider the following criteria when selecting a repository:
- Persistent identifiers, such as DOIs or PDB IDs
- Adequate metadata for retrieval and interpretation
- Open data policies
- Long-term availability, including rigorous curation and commitment to perpetual access
- Quality certification from trusted third parties
Sample dataset citation
[2] M. Partridge, "Spectra evolution during coating," figshare (2014) [retrieved 13 May 2015],
https://doi.org/10.6084/m9.figshare.1004612.
4. Citations to Code and Design Files
Code and design files used by researchers are often tied to proprietary formats such as SolidWorks or Matlab, and OSA cannot reasonably ensure robust vetting, archiving, or display of such material. Nevertheless, such files can be valuable to readers and may be included as "additional" material with a paper, provided that the files are relevant to the paper and are stored and cited according to OSA's guidelines.
Code and design files should be placed in the OSA figshare platform or in an open, archival repository as described for deposits of large datasets. The items should be cited as described above for large datasets, and first mentions should use the following convention to increase visibility to readers: ". . . as we show in Code 1 (Ref. [3])."
Sample code citation
[3] C. Rivers, "Epipy: Python tools for epidemiology," figshare (2014) [retrieved 13 May 2015],
http://doi.org/10.6084/m9.figshare.1005064.

